Fusoportal is an online database of Fusobacterium genomes sequenced by The Slade Lab at Virginia Tech using an Oxford Nanopore MinION and Illumina reads, and assembled using the software Unicycler developed by Ryan Wick in the group of Kat Holt. All genomes are annotated using Prodigal, and proteins are annotated using HMMER, InterPro, and Gene Ontology (GO) terms via Blast2GO. All data is made available as clickable and downloadable links to files found in the whole genome map or pages for individual genes. Why not take a look at the first full assembly of F. nucleatum 23726, a genetically tractable bacterium that is biomedically relevant in multiple diseases including colorectal cancer.
In addition, all raw data is available under NCBI BioProject PRJNA433545
The first genome we sequenced and provide here is Fusobacterium nucleatum subsp. nucleatum (strain ATCC 23726 / VPI 4351). We chose this genome because it is genetically tractable, but we found several limitations of previous genomes on the internet that were in 67 contigs, and contained breaks in several genes we were studying. We will be expanding FusoPortal to include more genomes in the near future. Access all available genomes here.
Want to screen one of your proteins to find out if there is a copy in a Fusobacterium genomes? We have implemented a custom FusoPortal BLAST server that will allow you to search one, two, or all of our genomes simultaneously. Results can be cross referenced against our whole genome maps, and alignments and fasta files for individual genes are available in downloadable formats.
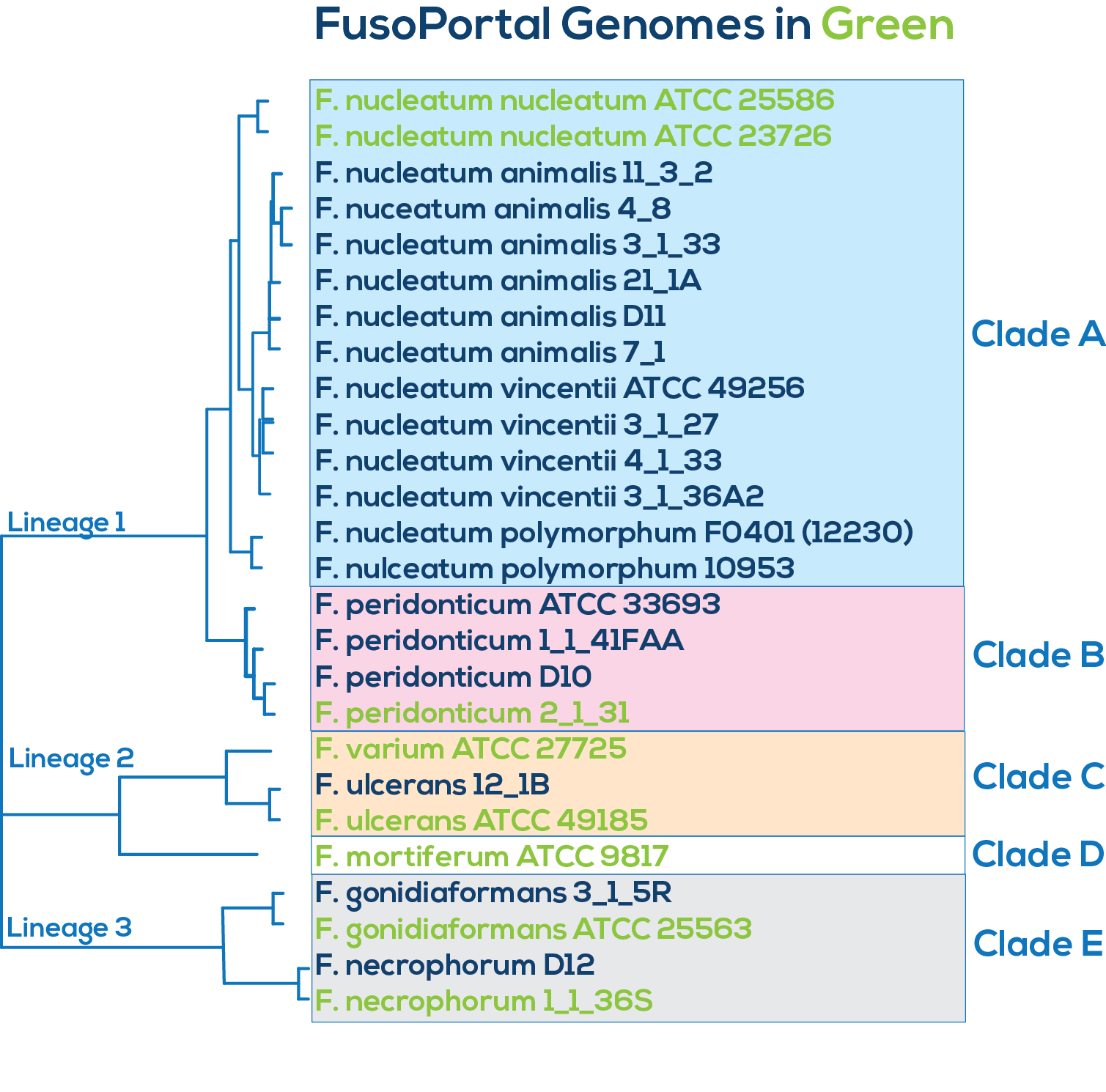 Here is a downloadable PDF of the tree above that contains links to each genome and sequence reads in the databases in which they were deposited. This tree was adapated from Manson McGuire et al, mBio 2014, from the laboratory of Ashlee Earl.
Here is a downloadable PDF of the tree above that contains links to each genome and sequence reads in the databases in which they were deposited. This tree was adapated from Manson McGuire et al, mBio 2014, from the laboratory of Ashlee Earl.
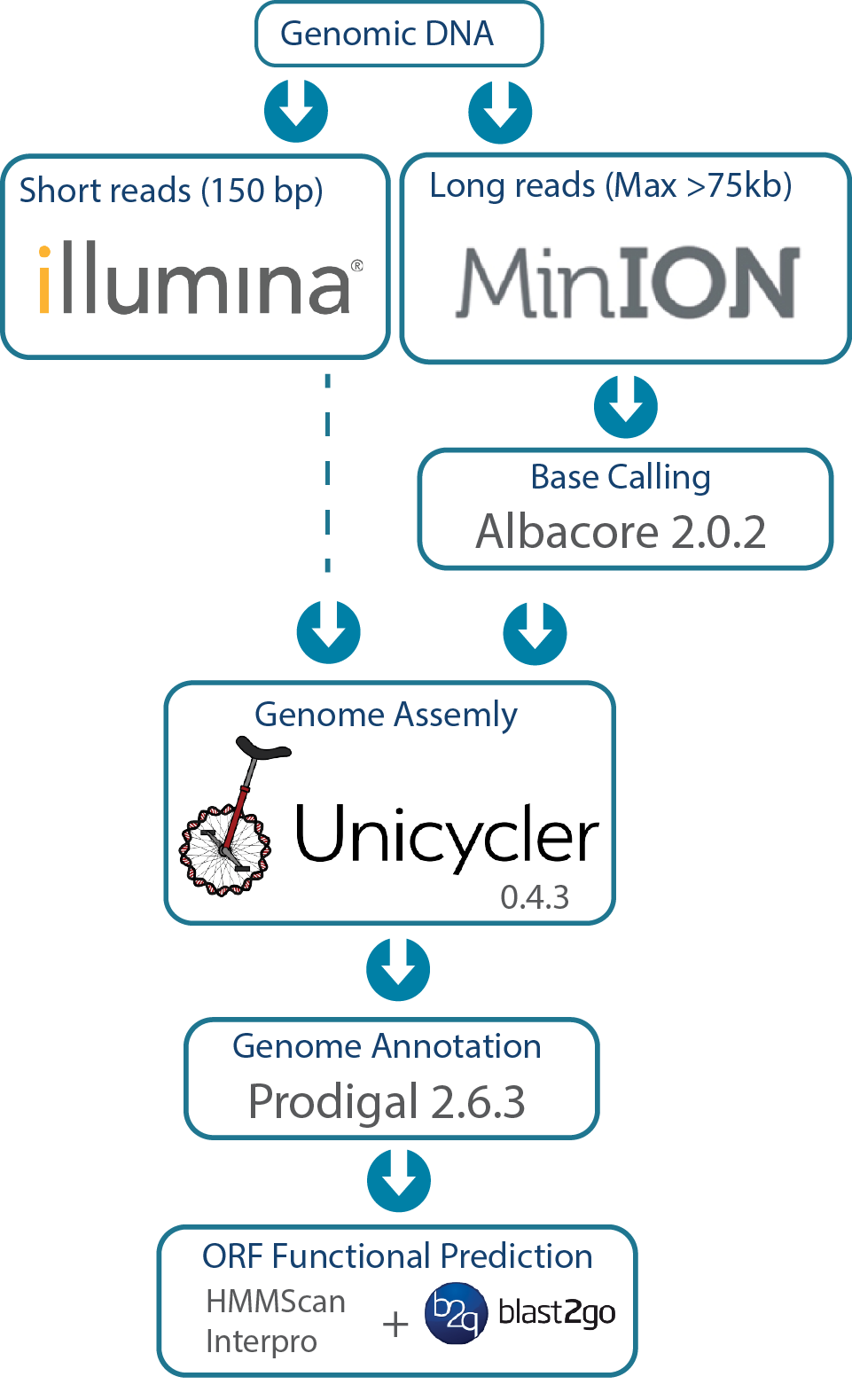
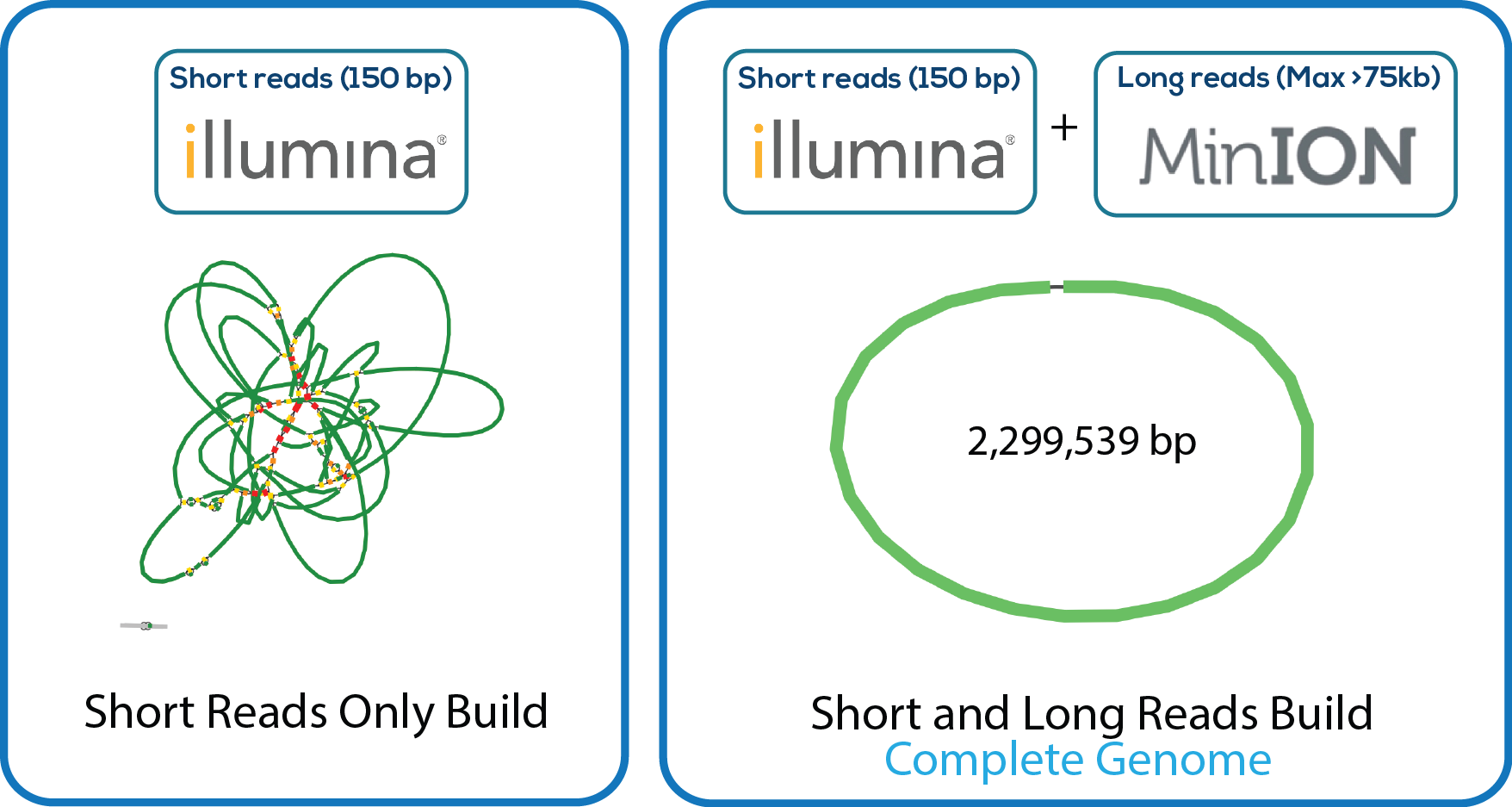
We used some amazing open source software by top developers. This pipleline is highly similar to that described by Wick et al. This robust system, which works well with multiplexing in both the MinION and Illumina runs, allows genomes to be sequenced, assembled, and annotated in house in a cost effective manner. We estimate that even with having the Illumina library prepped and sequenced at our core facility at Virginia Tech, the cost of each genome is <$500 US.
We built FusoPortal on the scalable HTML5 platform to give users a great experience across all platforms.
Members of the Slade Lab and beyond who helped build, expand, and maintain FusoPortal.