Check back to see when we have expanded with new genomes, or follow FusoPortal on Twitter
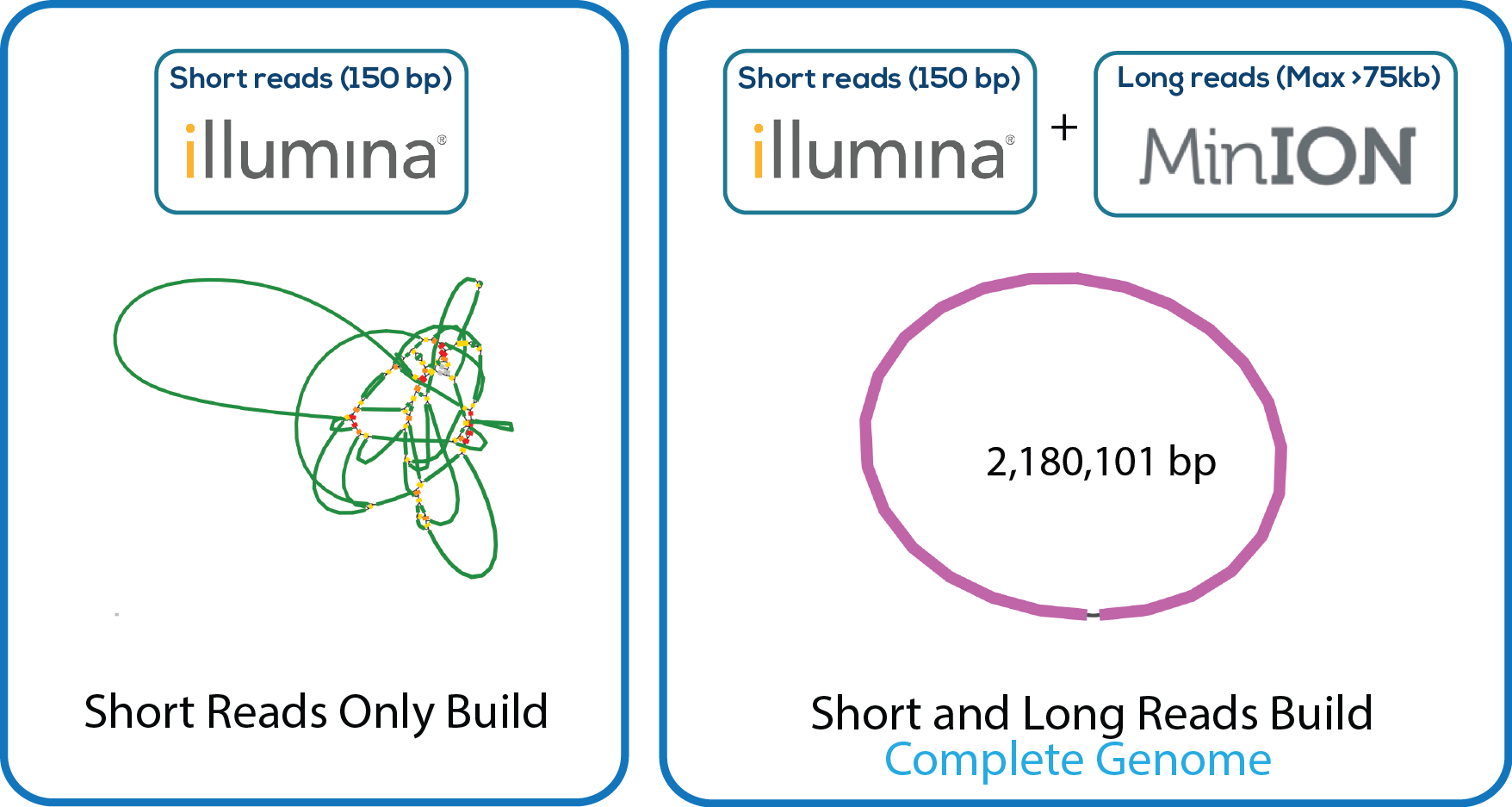
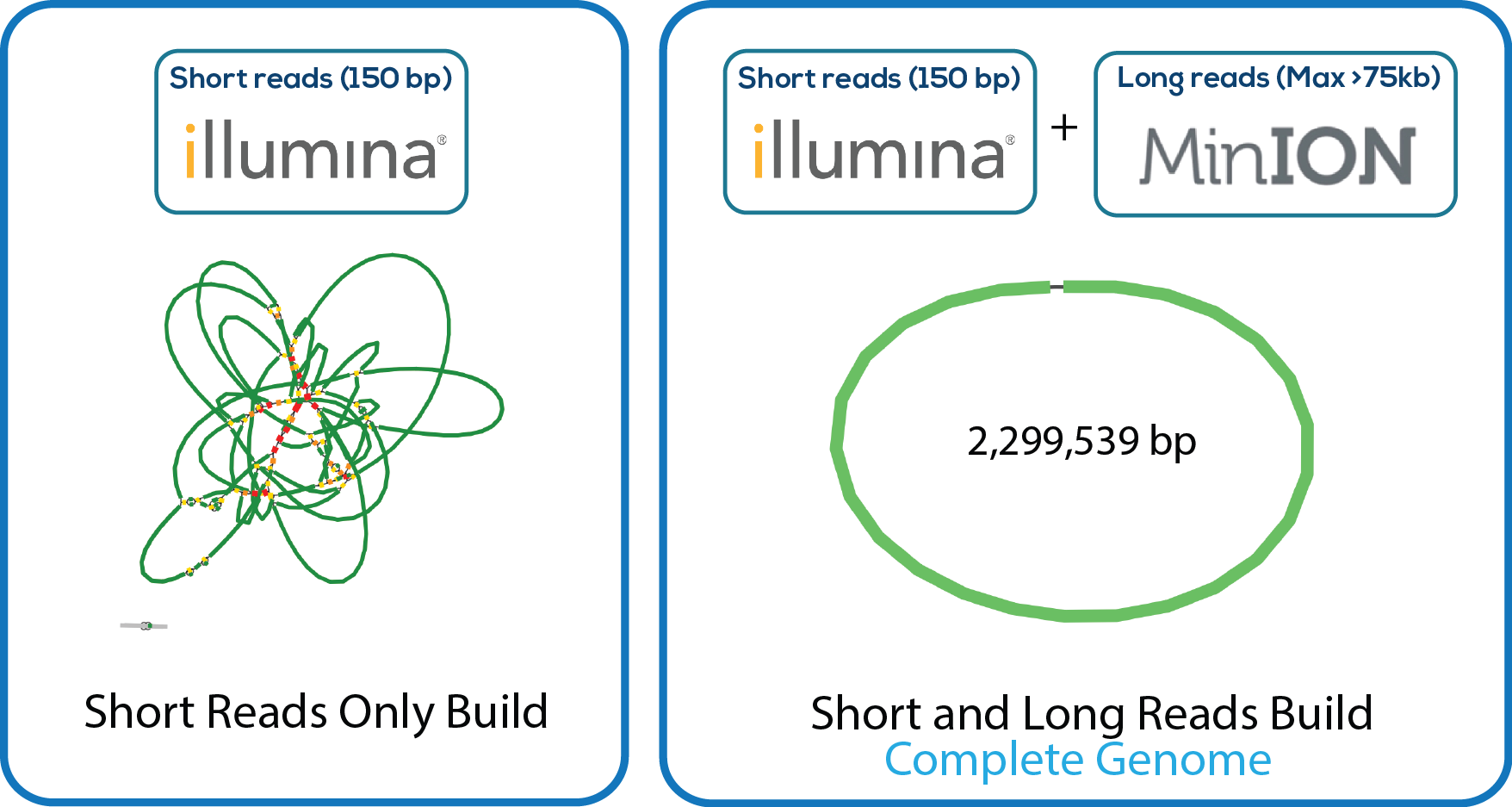
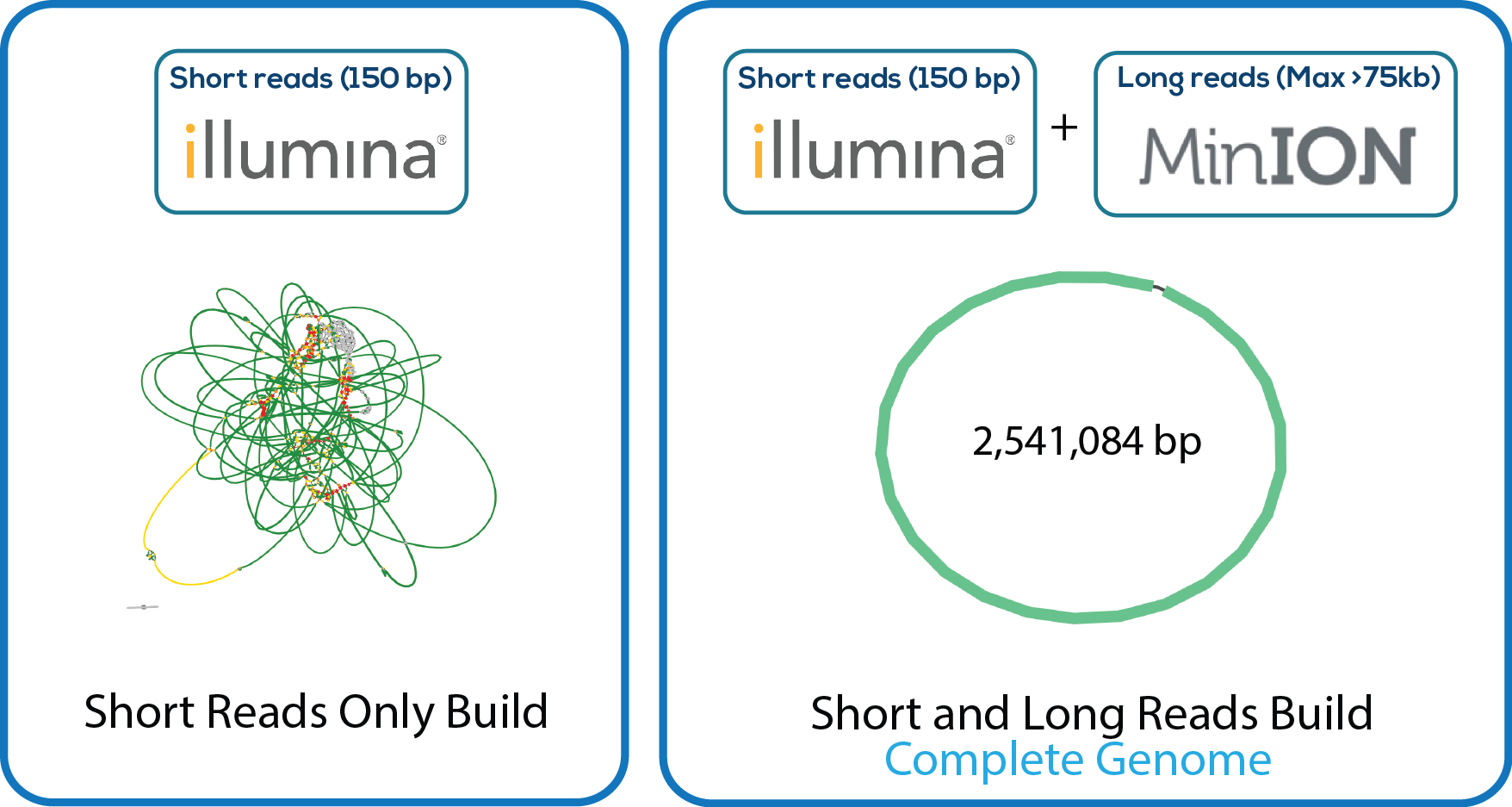
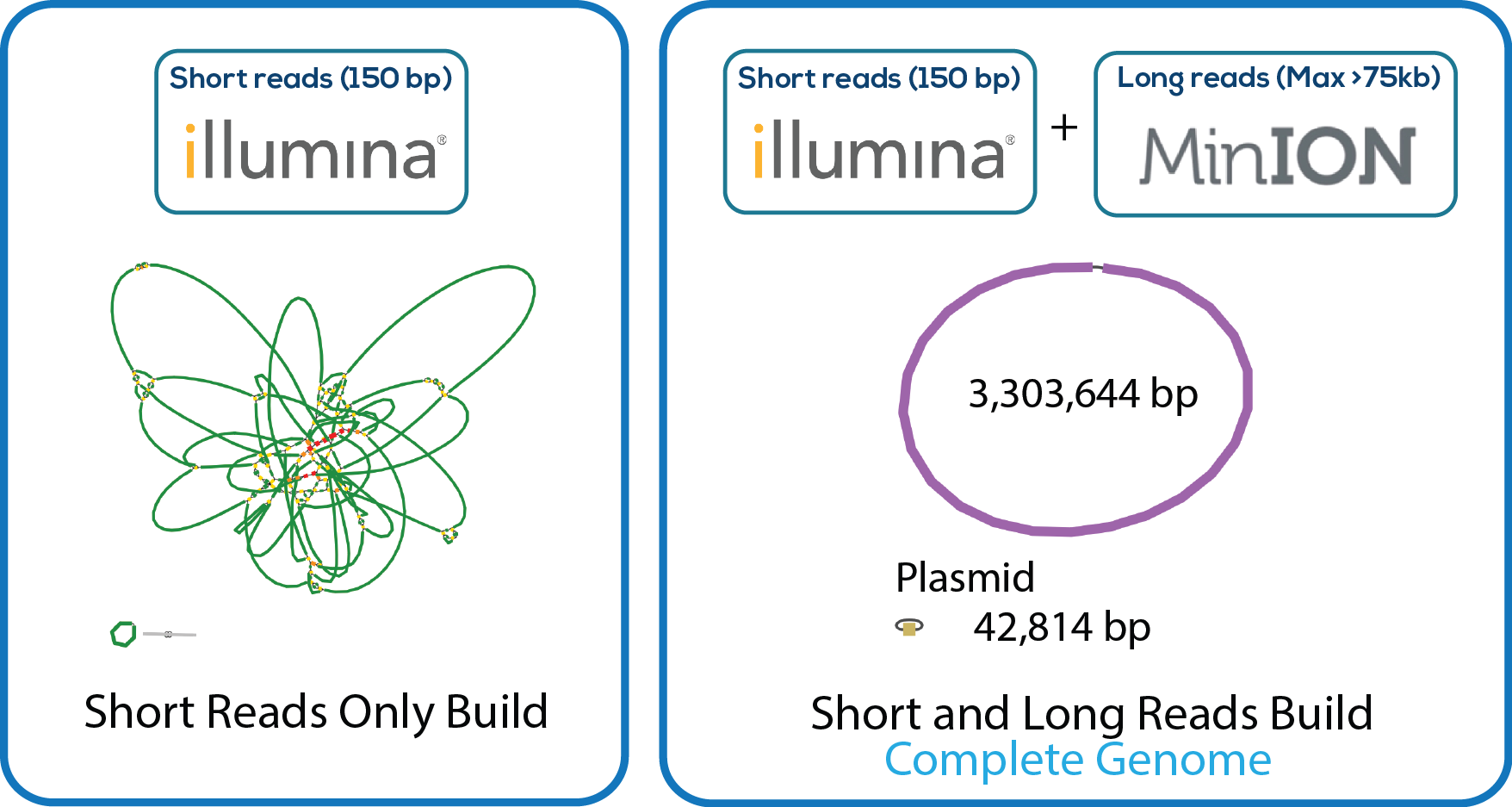
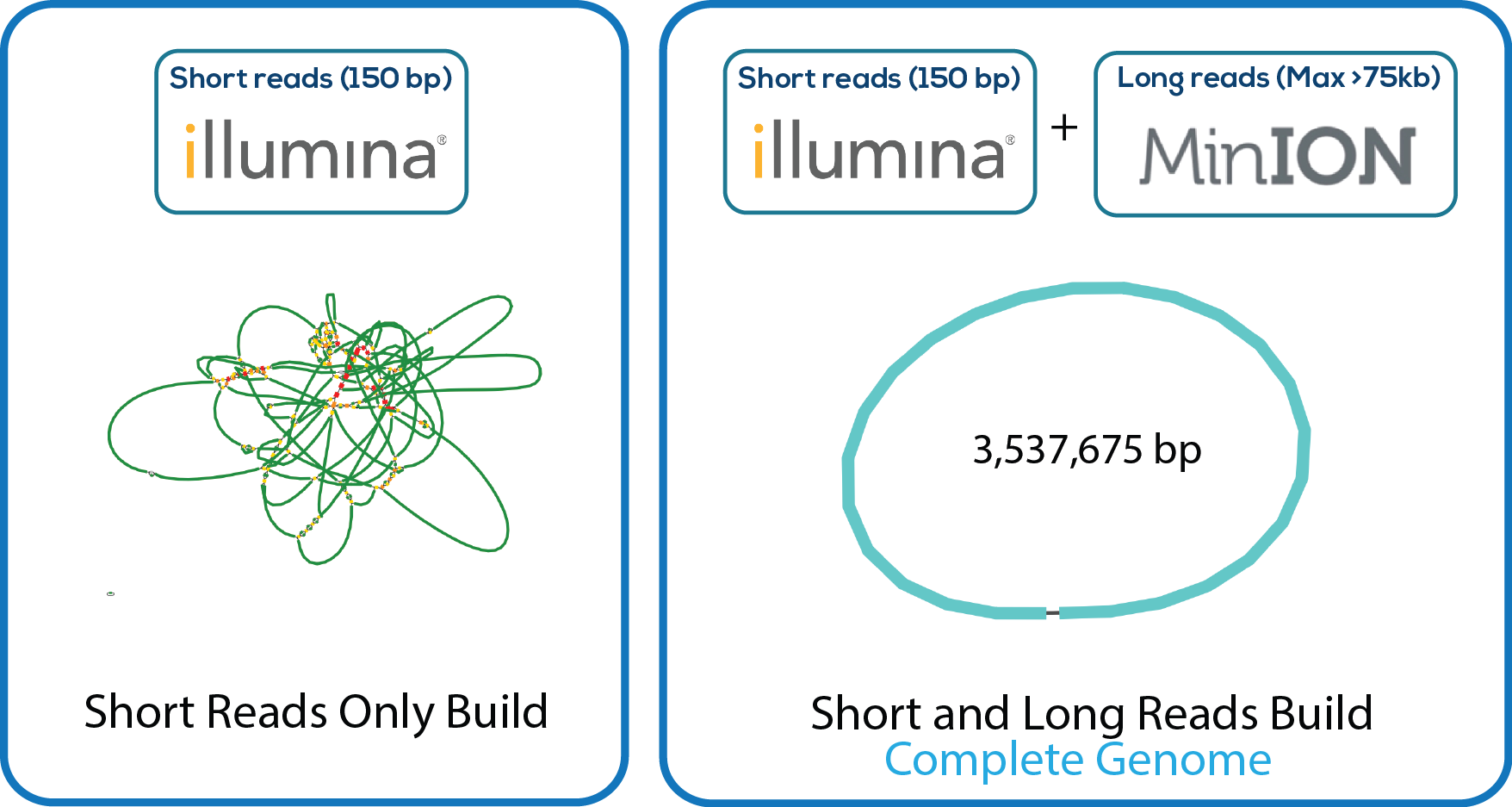
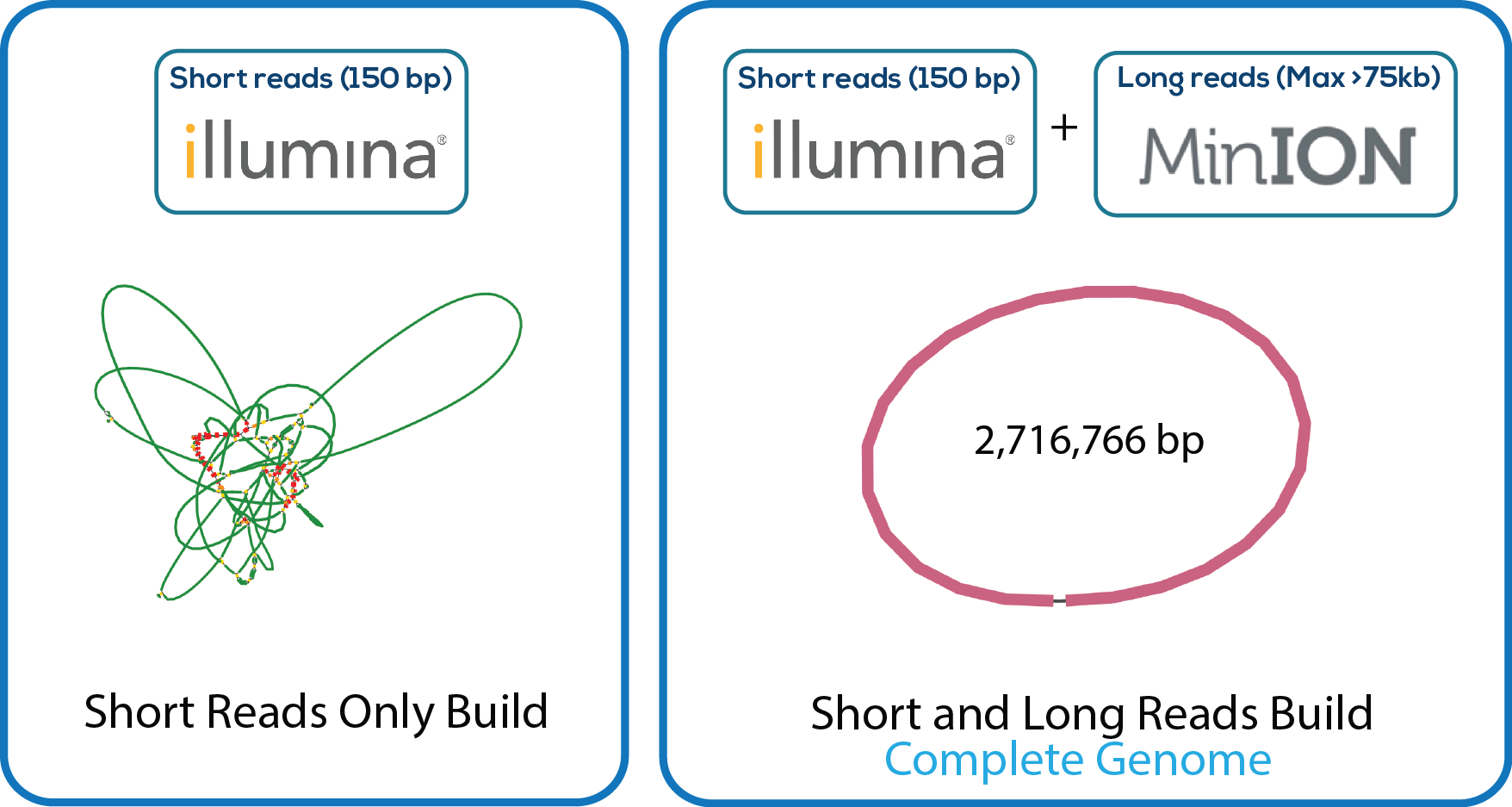
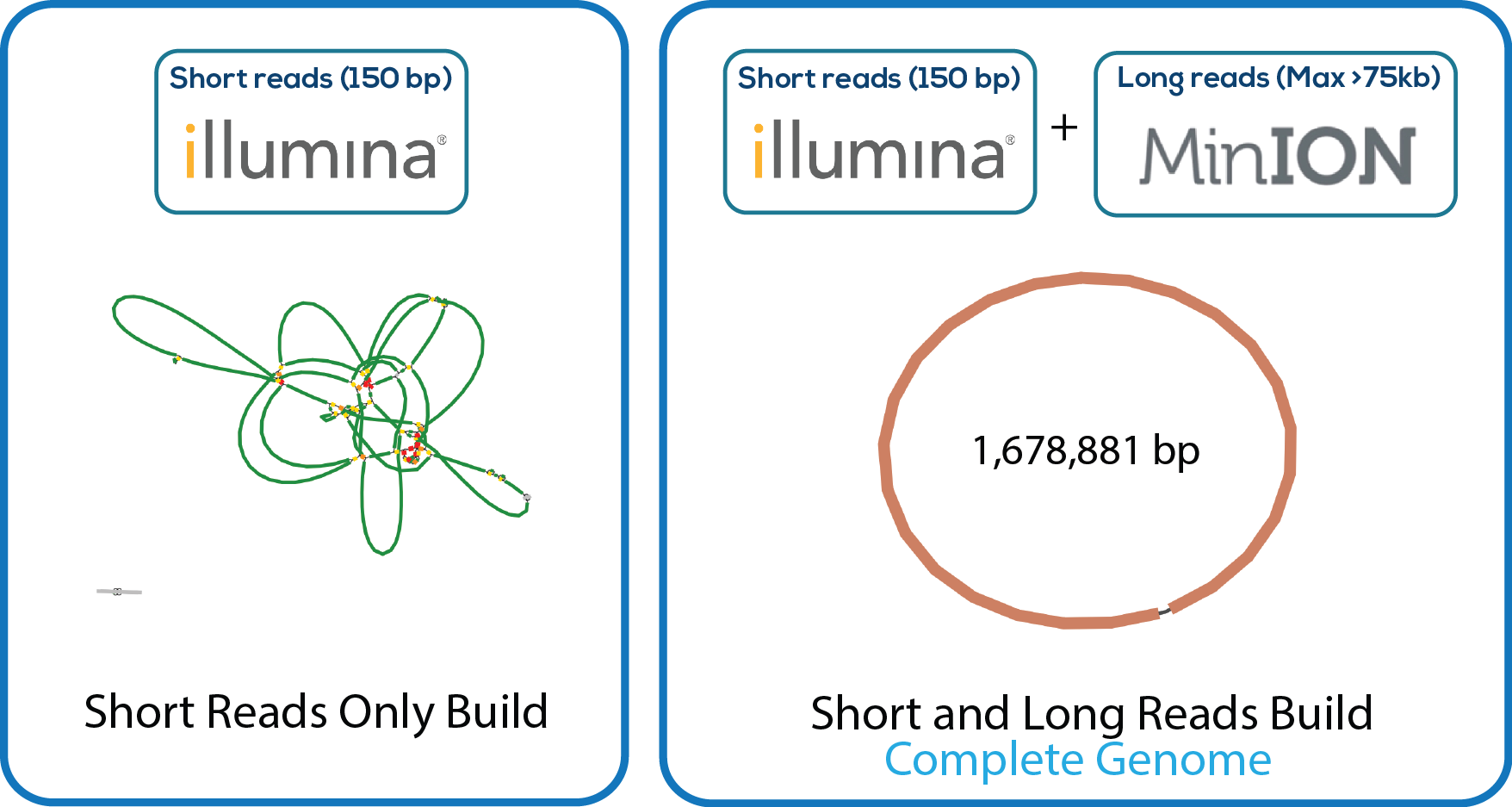
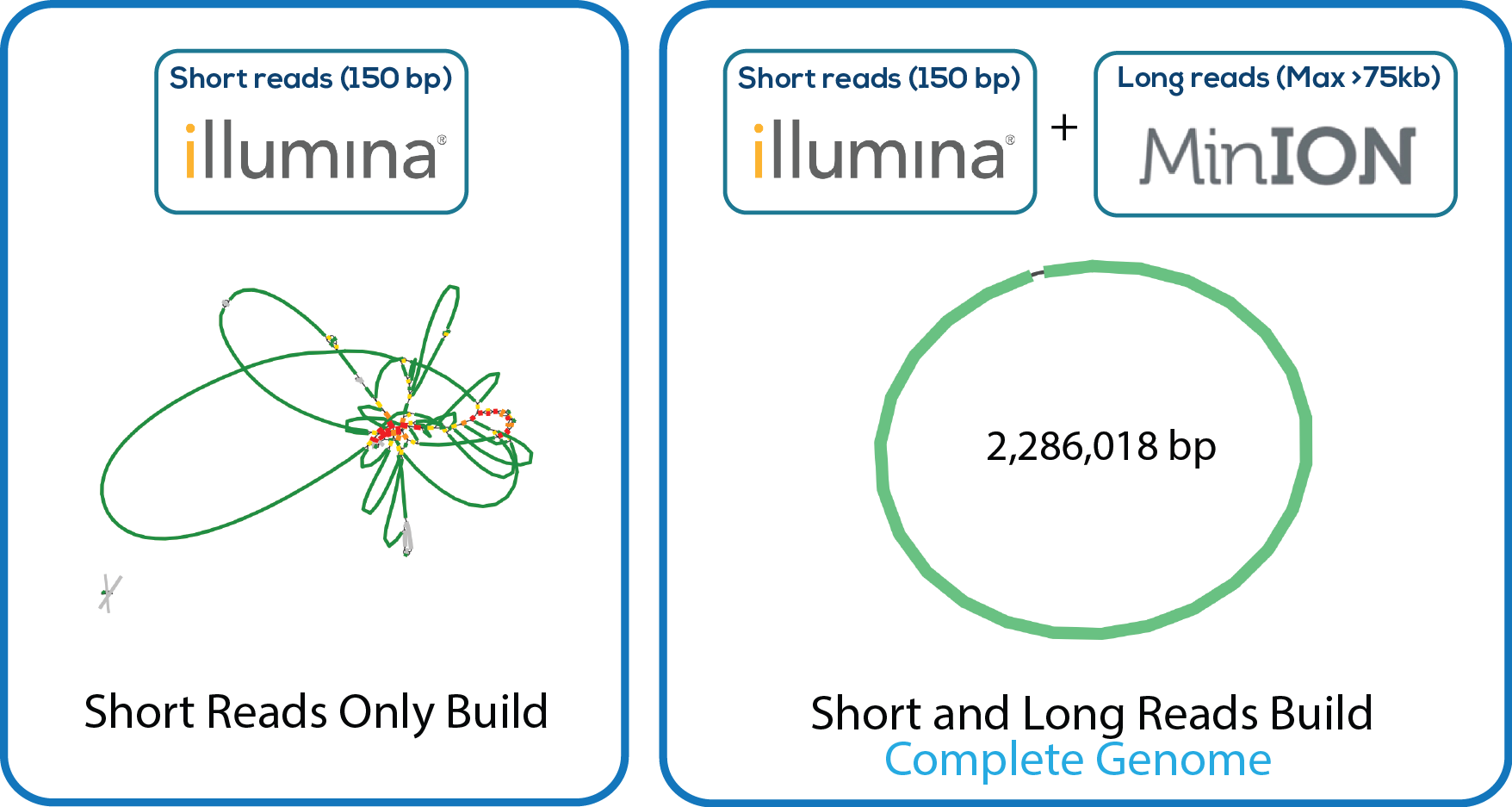
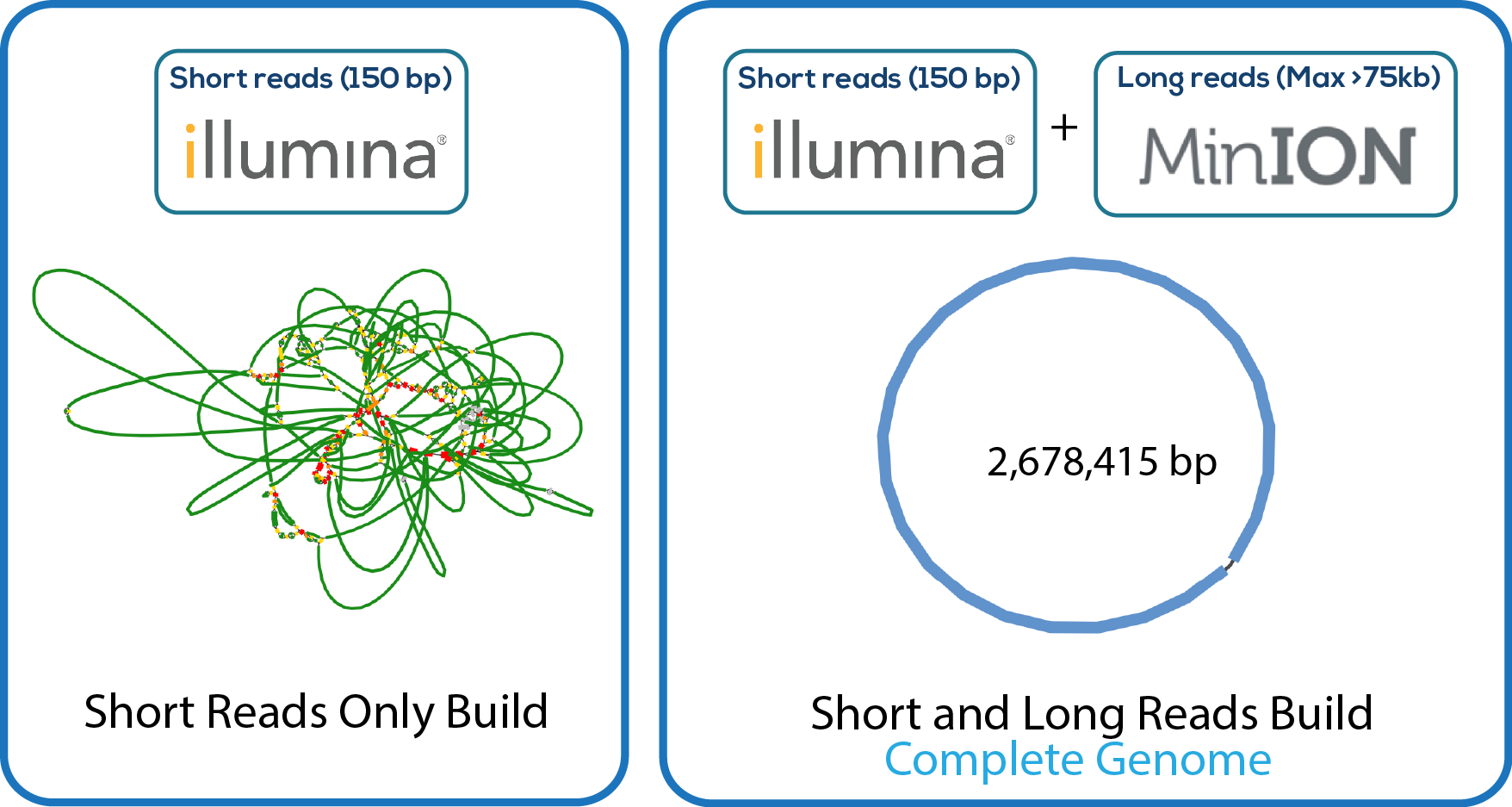
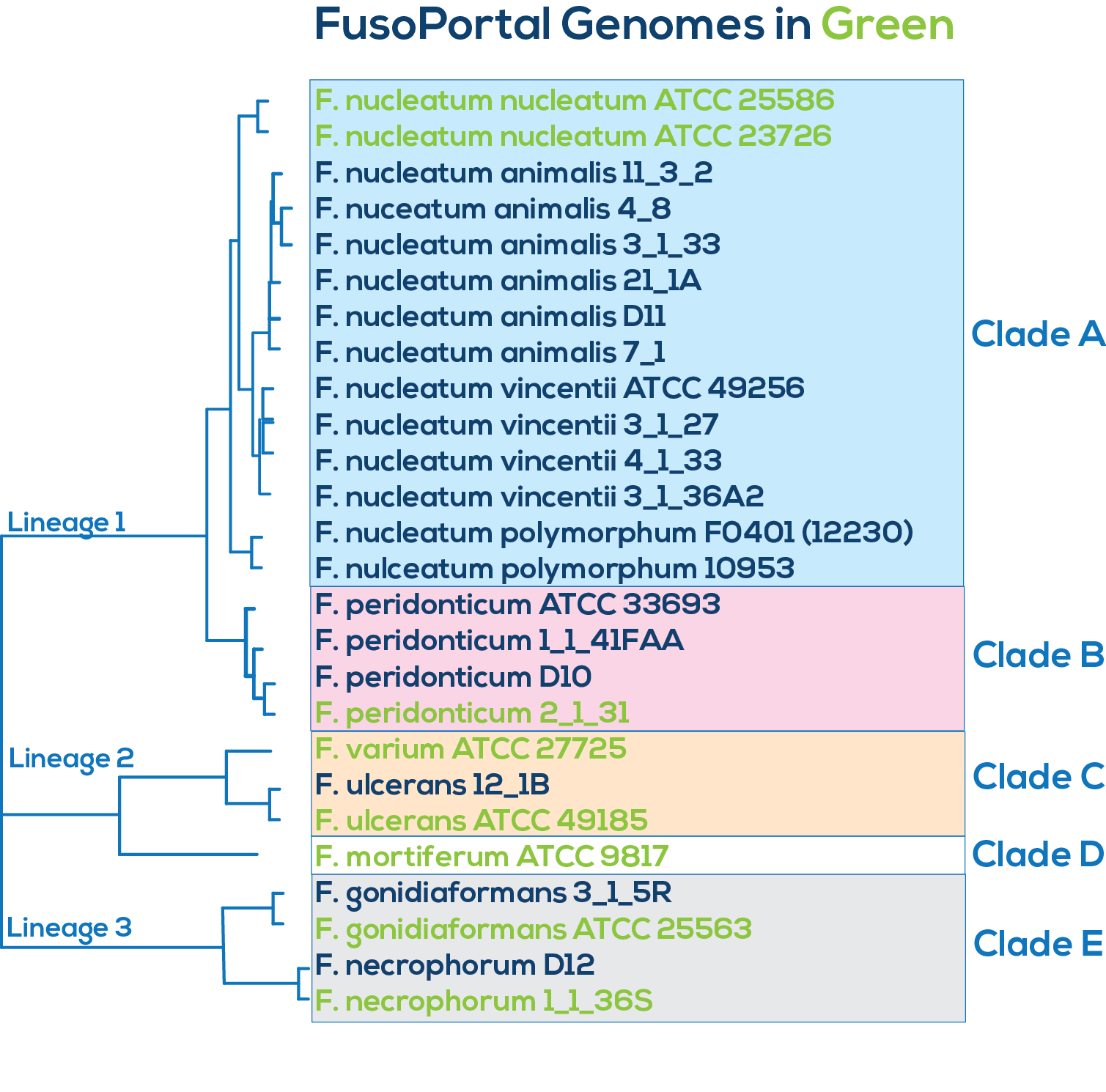 Here is a downloadable PDF of the tree above that contains links to each genome and sequence reads in the databases in which they were deposited. This tree was adapated from Manson McGuire et al, mBio 2014, from the laboratory of Ashlee Earl.
Here is a downloadable PDF of the tree above that contains links to each genome and sequence reads in the databases in which they were deposited. This tree was adapated from Manson McGuire et al, mBio 2014, from the laboratory of Ashlee Earl.